Publications
Teratology Primer, 3rd Edition
How Might Systems Biology Contribute to the Prediction of Teratogenic Risk?
Elaine M. Faustman, University of Washington, Seattle, Washington
Josh Robinson, University of California, San Francisco, California
Maxwell C. K. Leung, University of California, Davis, California
Development is a collection of interacting and dynamic processes that form an embryo, fetus, and ultimately a child. Much of what we understand has come about through study of isolated individual events occurring at an organ, cellular, or molecular level. However, in the dynamic events such as those that form an embryo, fetus, or a child, the interplay of individual events must also be understood. Systems biology seeks to study the relationships and interactions between various parts of a biological system (metabolic pathways, organelles, cells). This approach can contribute to our understanding of normal development and how it may be perturbed by a teratogenic exposure.
Teratologists think as systems biologists, either consciously or intuitively. In order to understand development, the conceptus is thought of as the maternal-child unit from the very start of life. While this approach is not unique to teratologists (many systems biologists and engineers model and understand processes as a whole), it is unique that teratologists have developed both disciplinary and scientific approaches that allow for such an integrated examination of normal and altered development.
Early embryologists used hierarchical and temporal approaches to understand the origin of tissues and cells. For example, the developing organism moves from the blastula to gastrula to neurula stages, and organs form from three tissues, ectoderm (neural plate, neural crest, and epidermis), mesoderm (dorsal –cephalic and trunk notochord and somites, ventral-blood islands and lateral plate organs including heart and kidney), and endoderm (yolk cells and alimentary canal organs such as lungs, liver, and stomach). Such tissue hierarchies are highly relevant for predicting impacts across species.
Systems biology provides a framework to follow the interconnectedness and dependencies of the different processes of development. Recent research has emphasized the importance of using cell, organ, and embryo cultures to understand the details of tissue and cell interactions. Only by looking at how these interactions build upon levels of biological complexity, moving from genetic and epigenetic, molecular, cellular, multicellular, tissues, organs, organ systems, to whole organisms can we understand overall development. There is genomic conservation so observations made at these levels are highly conserved and relevant across species. Examples include the relatively few (17) cell signaling pathways that have been characterized in all bilateral organisms that are able to explain most of development. For example, hedgehog signaling pathways, present in both vertebrate and Drosophila development, direct spermatogenesis in vertebrates and oogenesis in Drosophila. Hence, there is both a conserved but also a species-specific component that requires a systems approach in order to interpret impacts.
Recent advances in computational approaches have allowed systems biologists to become increasingly sophisticated in their ability to quantify impacts at one level for outcomes observed at more complex levels, birth and functional development (see chapter by Thomas Knudsen). In particular, such computational approaches have shown promise for answering more detailed questions about mode of action for teratogenic exposures, for improving cross-species extrapolation, for quantitative structure activity relationships, and for improving our understanding of gene-environmental interactions and responses. A systems biology approach also allows for evaluation at molecular, cellular, organ, conceptus, or population levels and can allow for better extrapolation across biological levels of observation. There has been tremendous progress made in the use of cell systems and organ culture to examine various effects on development. Linking the knowledge about the toxicokinetics and dynamics of chemical impacts has allowed for better prediction of potential for impacts at the organism level.
Two examples described below illustrate how a systems based approach can be used to evaluate developmental toxicology. A hallmark of such an approach is the use of data from different levels of biological complexity, form, and type. For example, knowledge about how a specific syndrome is defined phenotypically, and temporally expressed in both rodents as well as humans can be revealing.
Male developmental reproductive toxicology was investigated by M. Leung et al. 2016 using systems biology examples. These researchers conducted a review of rodent studies that evaluated male reproductive endpoints from studies that were identified in ToxRefDB, a comprehensive animal database from the US EPA. Endpoints included malformations, testicular atrophy, sperm effects, and tumors. Chemicals were identified with affected male developmental endpoints. Evaluation identified the lowest effect level at which the endpoint occurred with each system (mouse, rat, paternal, and offspring), experiment condition (time period and dose tested) and chemical tested (over 774 chemicals were included in the database at the time this work was done). Leung et al. identified prenatal development, sub-chronic and chronic, multigenerational reproductive and one-generational reproductive studies. in vitro assays were evaluated and identified from ToxCastDB (an EPA database of high-throughput in vitro assays) for bioactivities relevant for male reproductive events during development. This was accomplished by defining relevant assays as those that intersected with the in vivo database for male developmental endpoints of interest.
Twenty-three male reproductive endpoints were identified and classified into five types of categories relevant for male reproductive systems, including malformations, testicular tumors, sperm effects, and reproductive organ weight changes and histological changes. In this case study malformations were identified and included reduced anal-genital distance, hypospadias, cryptorchidism, and abnormal nipple retention, all of which were linked to early life-stage (prenatal, early postnatal) exposure scenarios. These databases allowed the investigators to look at endpoints across life-stages and in different generations, such as P1 (parental generation) versus F1 (filial generation). The comparisons allowed the investigators to identify chemicals that produced similar effects in all species or were species specific. They allowed for systematic comparison of species differences in sensitivity and by endpoint specificity Combinations of endpoints were also evaluated by chemical exposure and species.
In a similar fashion, evaluation of the in vitro systems allowed for the identification of results for specific receptor-mediated assays as well as cell growth and differentiation. Specific endocrine pathways and cellular and molecular changes such as vascularization and angiogenesis genes were identified as molecular targets. Of particular interest in this study were impacts on metabolism genes such as the cytochrome P450 specific genes for pathways that controlled synthesis as well as modification of hormones such as testosterone. A phenotypic hierarchy for testicular developmental pathways was identified for 54 chemicals.
In Leung et al. 2017, the authors used a systems biology approach to understand how stress and developmental pathways have evolved over time (Figure 1). This application shows that linking high-throughput data with evolutionary principles in a systems biology approach can identify conserved pathways across species. Ultimately, understanding species differences will improve adverse outcome pathway modeling and help toxicologists embrace variability in response.
Figure 1. Evolutionary origins of stress response and developmental pathways. Leung et al. 2017, reprinted with permission.
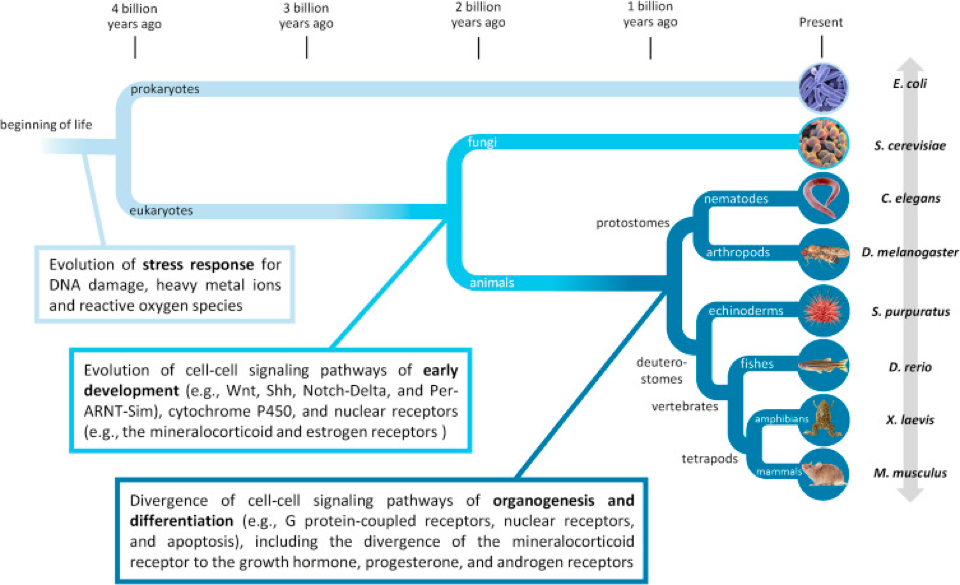
The availability of databases of compiled developmental studies that can be queried and in vitro assays allow this systems based approach to use common gene ontologies and common identifiers for chemicals (in this case over 774 chemicals) and common phenotypic profiles to accomplish these integrated evaluations. This is an example of systems biology application but also of the required data needs for establishing such evaluations. The toxicology programs at EPA are an excellent example of how these approaches can be integrated to answer important chemical and developmental endpoint questions.
A second example of using systems based approaches for addressing developmental toxicology questions can be seen in a set of papers by J. Robinson et al. These papers define how to address both genetic and environmental factors for developmental toxicology using systems biology for integration. This case study is with neural tube defects (NTD). NTDs occur at a rate of approximately 0.2 to 3.5 per 1000 births and are among the most common birth defect. Reviews of the genetic contribution to NTDs in humans and rodents were available and a candidate gene list was established. Genetic epidemiological studies were identified for human cases and quantitative trait loci analysis (QTL) allowed for the identification of genetic regions that conferred susceptibility for NTDs. Mouse knock-out studies and identification of genetically sensitive mouse strains, such as Swiss Webster Vancouver (SWV) inbred mice, were used along with genetic linkage assessments to identify the candidate genes in the rodent models, allowing these investigators to establish genome wide associations (GWAS) study databases. Using both sensitive and resistant mouse strains, SWV and C57BL6 mice, these investigators exposed the rodents to environmental chemicals (cadmium and methylmercury)that could cause neural tube defects. These environmental wide associate studies (EWAS) studies in rodents allowed for identification of candidate genes following exposure. By using a systems biology approach, these gene lists were integrated with toxicogenomic gene expression analysis and bio informatics tools including gene ontology databases that linked genes with specific gene pathways and function. Pathways were identified as common across humans and rodents with normal development as well as identifying pathways linked with NTDs across species. This case study illustrated how information from humans and rodents was collectively evaluated and both genetic and environmental factors contributing to the sensitivity to NTDs were identified.
An important implication of a systems biology approach is that in order to understand normal as well as altered development, teratologists are needed from diverse scientific and clinical disciplines. Clinicians such as obstetricians who are teratologists follow the course of pregnancy and may see birth defects early in gestation using ultrasound imaging. Dysmorphologists are trained to look at developmental processes and to diagnose syndromes and alterations in development that represent deviations from such processes resulting in malformations. Developmental biologists study details on the mechanisms of organ and tissue development. Molecular biologists look at comparable cellular and molecular processes in order to follow alterations that result in birth defects. Developmental toxicologists and pharmacologists study how chemicals or drugs can alter normal development and cause birth defects and developmental toxicity. An integration of knowledge from these many disciplines using the principles of systems biology will speed the understanding of teratogenic risk and an increased ability to minimize or prevent birth defects.
Suggested Reading
Boyles AL,Hammock P,Speer MC, et al. Candidate gene analysis in human neural tube defects. Am J Med Genet C Semin Med Genet 2005:135C, 9–23.
Copp AJ. Neurulation in the cranial region–normal and abnormal. J Anat 2005:207, 623–635.
Edwards SW, Preston RJ. Systems biology and mode of action based risk assessment. Toxicol Sci. 2008: 106(2):312–8. Epub 2008 Sep 12.
Faustman EM, Gohlke JM, Ponce RA, Lewandowski TA, Seeley MR, Whittaker SG and Griffith WC. Experimental Approaches to Evaluate Mechanisms of Developmental Toxicity in developmental and Reproductive Toxicology: A Practical Approach. Hood RD, (ed.) Hoboken, N.J., CRC Press, Taylor and Francis Group 2006.
Gohlke JM, Griffith WC and Faustman EM. A systems-based computational model for dose-response comparisons of two mode of action hypotheses for ethanol-induced eurodevelopmental toxicity. Toxicological Sciences. 2005; 86(2):470–484.
Gohlke JM, Griffith WC and Faustman EM. Computational models of neocortical neuronogenesis and programmed cell death in the developing mouse, monkey, and human. Cerebral Cortex. 2007; 17(10):2433–2442.
Harris MJ, Juriloff DM. Mouse mutants with neural tube closure defects and their role in understanding human neural tube defects. Birth Defects Res A Clin Mol Teratol. 2007, 79: 187–210.
Ideker T, Galitski T, and Hood L. A new approach to decoding life: Systems biology. Annu. Rev. Genomics Hum. Genet. 2001:2, 343–372.
Kavlock RJ, Ankley G, Blancato J, Breen M, Conolly R, Dix D, Houck K, Hubal E, Judson R, Rabinowitz J, et al. Computational toxicology a state of the science mini review. Toxicol. Sci. 2007:103, 14–27.
Kitano, H. Systems biology: A brief overview. Science 2002: 295, 1662–1664.
Knudsen TB, and Kavlock RJ. Comparative Bioinformatics and Computational Toxicology. In Developmental Toxicology, 3 edition. B. Abbot and D. Hansen (Eds.). pp. 311–360. Taylor and Francis. (in press) 2008.
Leung MCK, Procter AC, Goldstone JV, Foox J, DeSalle R, Mattingly CJ, Siddall ME, Timme-Laragy AR. Applying evolutionary genetics to toxicology and risk assessment. Reproductive Toxicology. 2017:69, 174-186
Leung CL, Phuong J, Baker NC, Sipes NS, Klinefelter GR, Martin MT, McLaurin KW, Setzer RW, Darney SP, Judson RS, Knudsen TB. Systems toxicology of male reproductive development: profiling 774 chemicals for molecular targets and adverse outcomes. Environmental Health Perspectives. 2016:124
National Research Council (U.S.). Committee on Developmental Toxicology and National Research Council (U.S.). Commission on Life Sciences. Scientific frontiers in developmental toxicology and risk assessment. Faustman E.M. (Chair) and Gerhart J. (Vice Chair). Washington, DC, National Academy Press. 2000.
Robinson JF, Port JA, Yu X, Faustman EM. Integrating genetic and toxicogenomic information for determining underlying susceptibility to developmental disorders. Birth Defects Research Part A: Clinical and Molecular Teratology. 2010:88, 920-930.
Robinson JF, Guerrette Z, Yu X, Hong S, Faustman EM. A systems-based approach to investigate dose-and time-dependent methylmercury induced gene expression response in C57BL/6 mouse embryos undergoing neurulation. Birth Detects Research Part B: Developmental and Reproductive Toxicology. 2010:89 (3), 188-200.
Slack JMW. From Egg to Embryo: Regional Specification in Early Development, 2nd edition. Cambridge University Press, 1991, p.348.
